Discover our new platform: Learn more about spatialx
 An
analytics hub for public single-cell and spatial data, built upon a comprehensive
database, providing an innovative approach for in-depth research at a large scale.
An
analytics hub for public single-cell and spatial data, built upon a comprehensive
database, providing an innovative approach for in-depth research at a large scale.

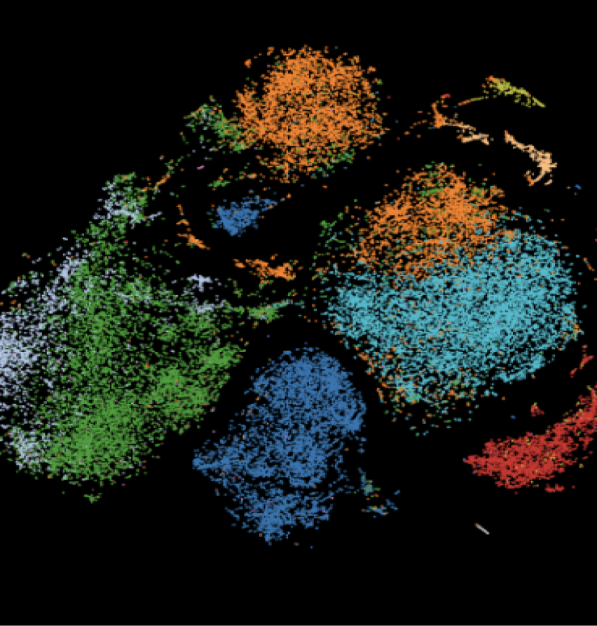
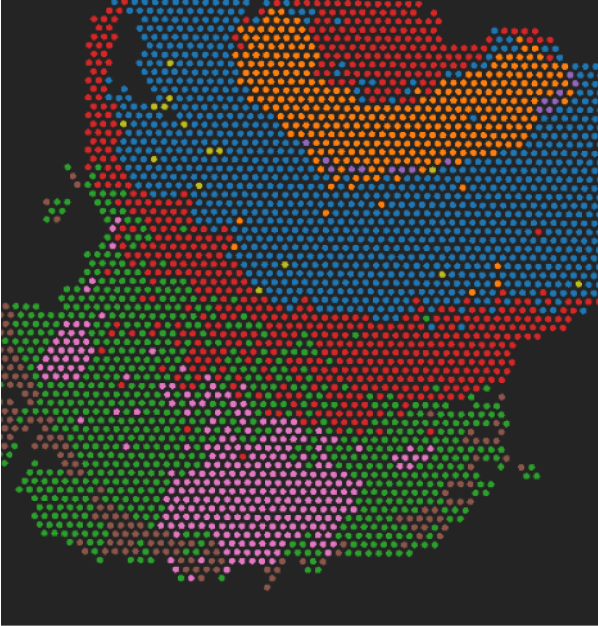
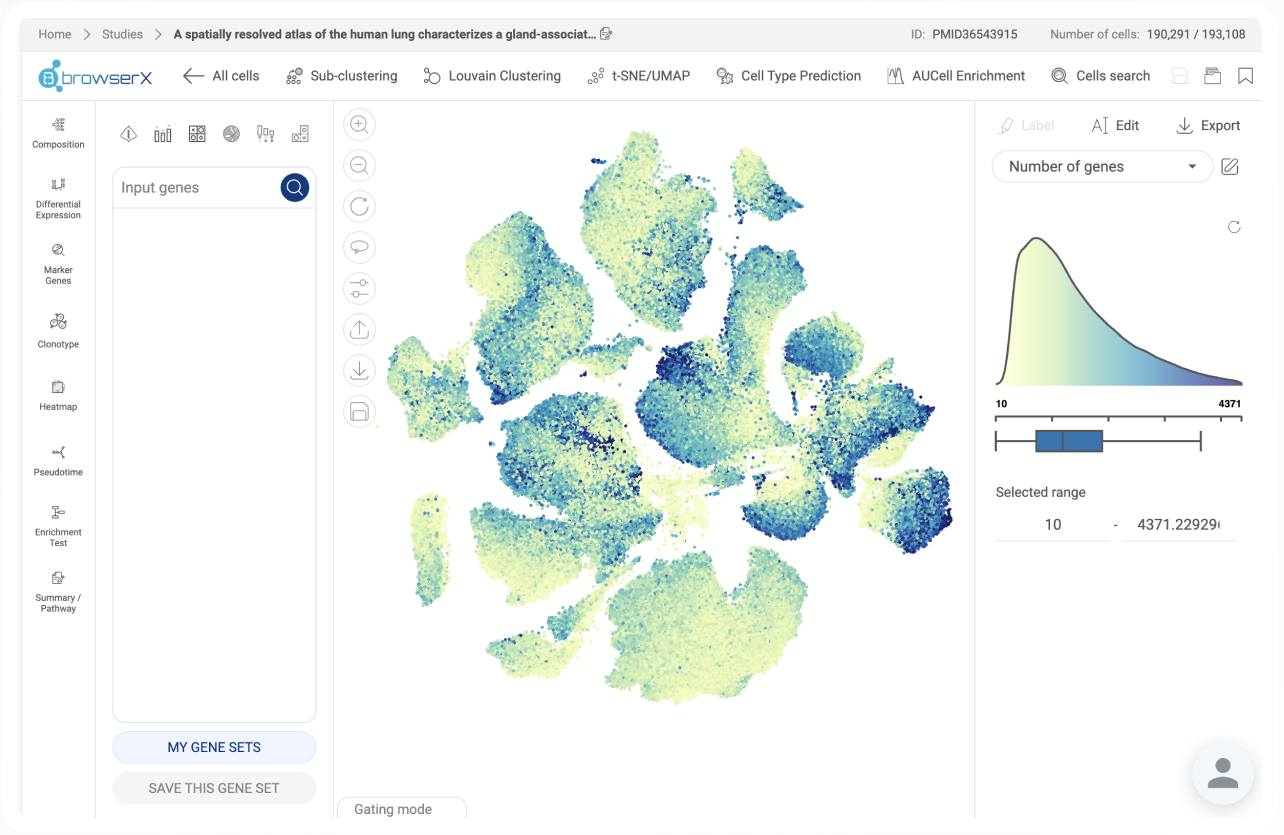
“BioTuring BBrowserX Pro delivers high-quality t-SNE visualizations for analyzing single-cell RNA-seq data from platforms such as 10x Genomics and Parse. The software also streamlines the process of uploading and analyzing published datasets from NCBI. Users benefit from highly responsive and proactive customer support. Overall, the BioTuring platform is a strong choice for scientists seeking fast and efficient single-cell data analysis.”
Slava Furtak
Argentys Informatics
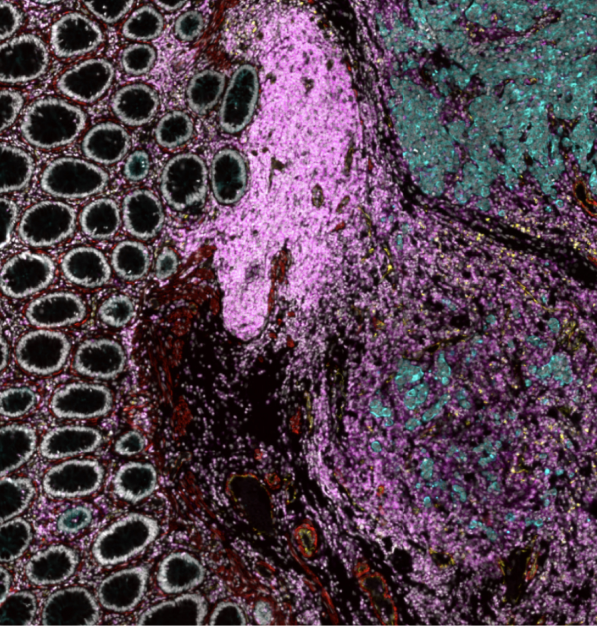
“BioTuring SpatialX is designed to explore enormous spatial data to advance scientific discovery through integrated algorithms and innovative software solutions. The unique Windows-based & non-coding platform allows us to tap the full potential of spatial biology. Its outstanding flexibility, versatility, and intuitive user interface makes it the perfect tool to analyze multidimensional datasets generated by various technologies. We as biologists can quickly interrogate our spatial data and better understand complex mechanisms of cell-to-cell interaction at single cell transcriptomics level, with superior BioTuring customer support.”
Xuhuai Ji, PhD & MD
Genomics Director
Human Immune Monitoring Center
Stanford Medicine
“BioTuring is fantastic! It has become an essential part of the toolkit for wet lab researchers who want to dive deep into single-cell transcriptomics. Its intuitive interface and powerful capabilities make data exploration both efficient and enjoyable.”
Di Zhao
Assistant Professor. UT MD Anderson Cancer Center
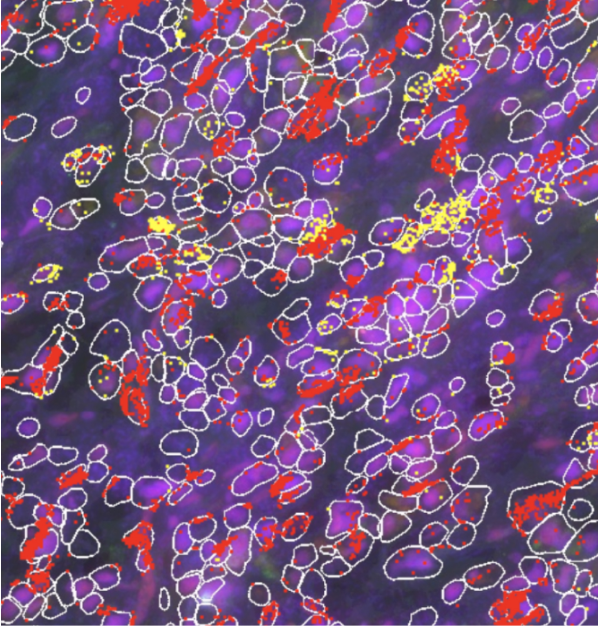
“SpaitialX is indispensable for visualizing our CosMx spatial data. It allows us to interpret results within the context of tissue architecture and neighboring cells. The user-friendly design helps researchers quickly extract meaningful biological insights.”
Daisuke Komura
Assistant Professor, The University of Tokyo
“BBrowserX Pro is an outstanding tool for single-cell transcriptome and TCR repertoire analysis. It’s fast, easy to use—even for beginners—and the technical support team has been incredibly responsive. It’s now a core part of our lab’s workflow.”
Motoko Kimura
Professor, Chiba University
“SpatialX offers a seamless solution for spatial transcriptomic analysis. Its powerful features and clean interface let us uncover insights without needing to code in R. It saves us time and effort while delivering publication-quality results.”
Jonathan Yap PhD.
Postdoctoral Fellow University of Hawaii, John A. Burns School of Medicine
“I originally learned single-cell analysis in R, but BBrowserX Pro has completely changed how I work. It makes interpreting data more intuitive and generates insightful visualizations with ease. It’s perfect for both learning and research.”
Ryan Lingerak
PhD Candidate CWRU, Bingcheng Wang Lab